use cases

MGnify oriented implementation for the Marine Genomic Observatories oriented pipeline, developed in the framework of an EOSC-Life funded project
metaGOflow is a pipeline based on MGnify and its workflows, aiming to address the challenges of the analysis of the European Marine Omics Biodiversity Observation Network (EMO BON) data. EMO BON is a long-term omics observatory of marine biodiversity that generates hundreds of metagenomic samples periodically from a range of stations around Europe.
In the following diagram is an overview of the metaGOflow steps:
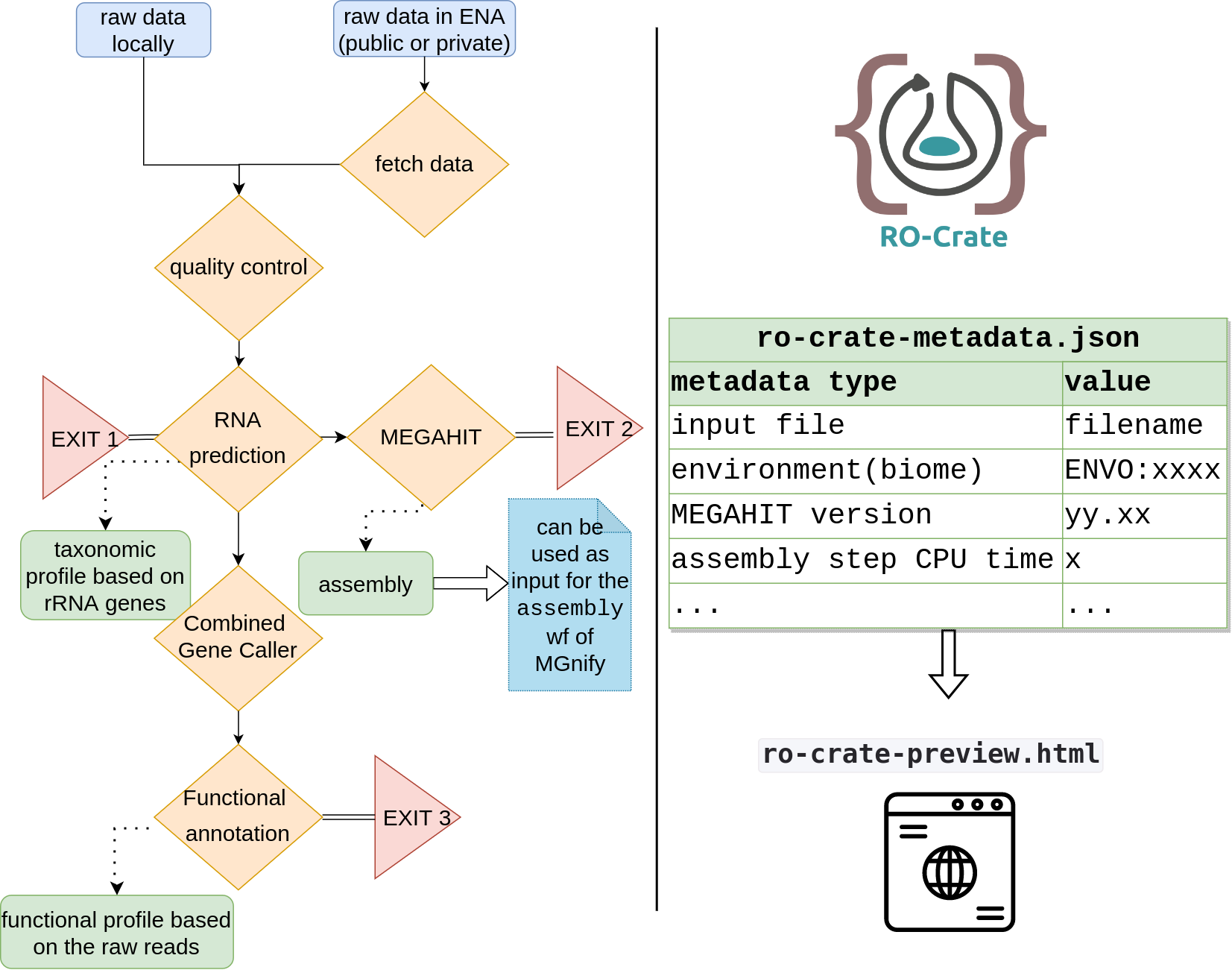
Here we show visual components accompanying the metaGOflow publication. We performed all steps of metaGOflow for an EMO BON marine sediment (ERS14961254) and a water column (ERS14961281) sample. A quality control report, the taxonomic inventories as well as some of the functional annotations returned in each case are displayed below.
- To have an overview on the metaGOflow results for the marine sediment sample click here.
- For the water column sample, you may have a look here
We also tested metaGOflow with a TARA OCEAN sample.
You may find the RO-crate produced from the complete metaGOflow run for this sample through this Zenodo repository.
Last, you may have a look on the ro-crate-metadata.json file that describes the RO-Crate metaGOflow built for this sample here.
metaGOflow source code: GitHub repo
Citation
In case you are using metaGOflow for your analysis, please remember to cite us:
metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data (to be submitted in GigaScience)
Contact
![]()